Targeted Variant Testing
Targeted Variant Testing (TVT) may be used in the following situations:
- Confirmatory analysis of research or results reported by another laboratory
- Testing of published founder or common variants
- Clarification of variant classification for variants reported by another laboratory
With Targeted Variant Testing, you can order up to 10 variants (see eligibility information in About Targeted Variant Testing) per order for a fixed price.
Targeted variant testing is limited to reporting only the presence or absence of the specifically requested variant(s) in the individual being tested. Incidental findings will not be reported, however, a recommendation for additional testing may be included on the report if incidental findings are present. See test information section below for additional information.
Summary
The Blueprint Genetics Targeted Variant Testing (test code TVT001):
Read about our accreditations, certifications and CE-marked IVD medical devices here.
Sample Requirements
- Blood (min. 1ml) in an EDTA tube
- Extracted DNA, min. 2 μg in TE buffer or equivalent
- Saliva (Please see Sample Requirements for accepted saliva kits)
Label the sample tube with your patient’s name, date of birth and the date of sample collection.
We do not accept DNA samples isolated from formalin-fixed paraffin-embedded (FFPE) tissue. In addition, if the patient is affected with a hematological malignancy, DNA extracted from a non-hematological source (e.g. skin fibroblasts) is strongly recommended.
Please note that, in rare cases, mitochondrial genome (mtDNA) variants may not be detectable in blood or saliva in which case DNA extracted from post-mitotic tissue such as skeletal muscle may be a better option.
Read more about our sample requirements here.
We do not accept prenatal samples from ongoing pregnancies for Targeted Variant Testing.
Please contact your local genetic services consultant or our Customer Support team support@blueprintgenetics.com to assist with ordering. We are here to help you!
Targeted Variant Testing may be used in the following situations:
- Confirmatory analysis of research or results reported by another laboratory
- Testing of published founder or common variants
- Clarification of variant classification for variants reported by another laboratory
With Targeted Variant Testing, you can order up to 10 variants per order for a fixed price.
Blueprint Genetics will only report the variant(s) of interest.
Eligibility
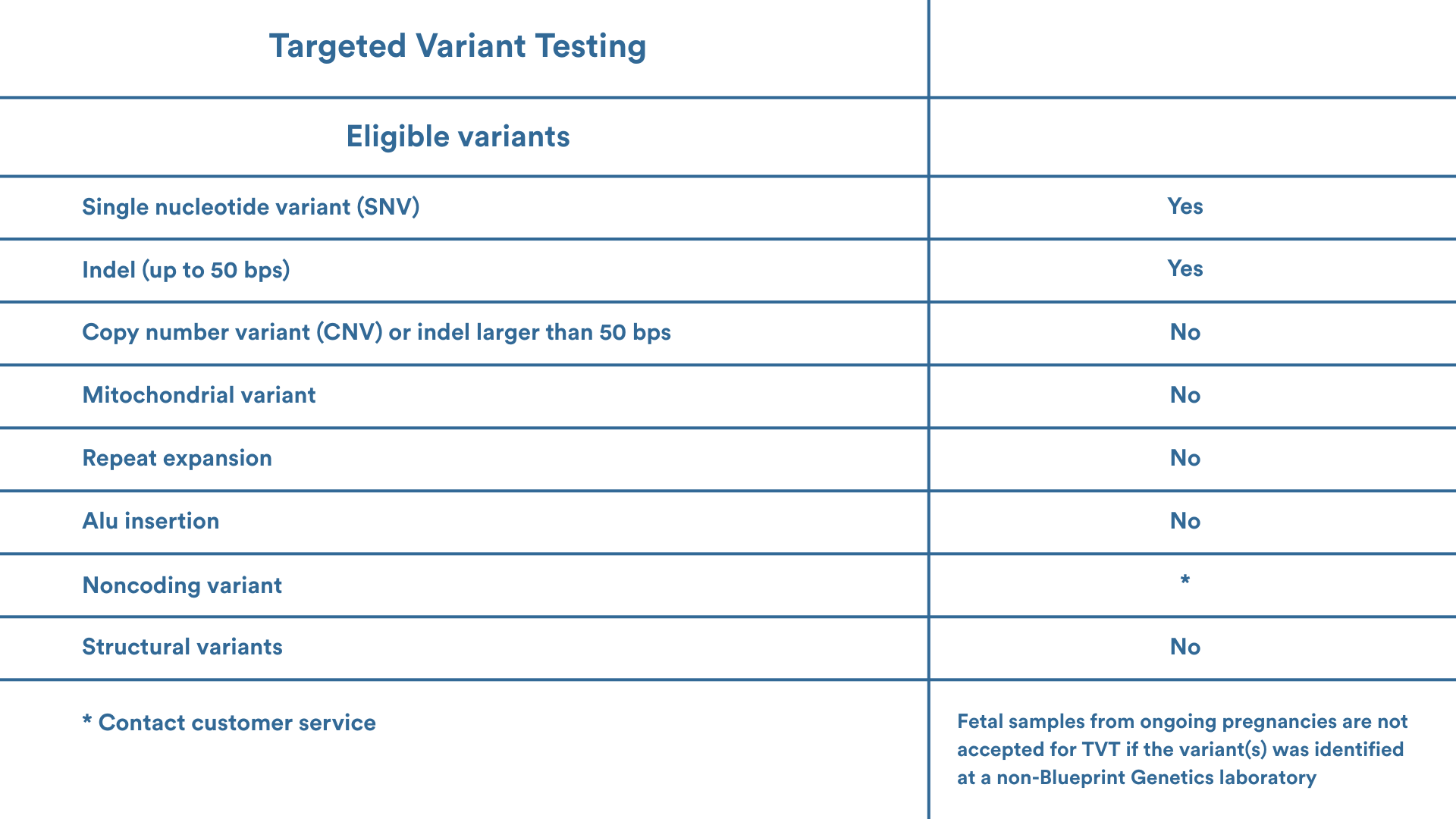
Please contact your local genetic services consultant or our Customer Support team support@blueprintgenetics.com to assist with ordering. We are here to help you!
How to order
- Log in to our online portal Nucleus at nucleus.blueprintgenetics.com for the easiest way to order and access results.
- Fill out both the consent and requisition forms.
- Send the sample to Blueprint Genetics.
- Sample Requirements:
- Blood (min. 1ml) in an EDTA tube
- Extracted DNA, min. 2 μg in TE buffer or equivalent
- Saliva (Oragene DNA OG-500 kit/OGD-500 or OG-575 & OGD-575)
- Read more about our sample requirements
- Sending a positive control sample from the index patient is not required.
- Sample Requirements:
Methods
Targeted Variant Testing is based on next generation sequencing (NGS) technology. Sanger sequencing or qPCR/digital PCR may be used as an orthogonal method as needed.
Targeted variant testing is limited to reporting only the presence or absence of the specifically requested variant(s) in the patient; however, other variants in the gene of interest may be visible to the interpretation team. If incidental findings are identified, the report may include a recommendation that additional testing be performed.
Test Strengths and Limitations
Test Strengths
- CLIA-certified personnel performing clinical testing in a CLIA-certified laboratory
- Powerful sequencing technologies, advanced target enrichment methods, and precision bioinformatics pipelines ensure superior analytical performance
- Comprehensive clinical statement
Test limitations
See eligibility information in About Targeted Variant Testing.
This test may not reliably detect the following:
- Low-level mosaicism
- Variants within pseudogene regions/duplicated segments
The genes on the panels have been carefully selected based on scientific literature, mutation databases, and our experience.
The panels are sectioned from our high-quality, clinical grade NGS assay. The panel analysis includes a combination of both sequence variants (single nucleotide variants (SNV’s) and indels) as well as deletions and duplications (copy number variants (CNV)).
Please refer to the table below for performance metrics of the analytical validation of the assay. The validation includes the evaluation of reference samples to determine the capability of the assay to detect various types of variants. The sensitivity values quoted in the analytic validation may not precisely reflect the performance in a production setting and is not a guarantee of the assay’s clinical performance. The provided performance metrics are based on a validation conducted at our laboratory in Finland. The assay has been validated for various sample types including EDTA-blood, isolated DNA (excluding from formalin fixed paraffin embedded tissue), saliva, and dried blood spots (filter paper cards).
Performance of Blueprint Genetics high-quality, clinical grade NGS sequencing assay for panels.
Analytical sensitivity to detect single-nucleotide variants and indels were calculated using both versions v3.3.2 and v4.2.1 of high-confidence region benchmark data provided by Genome in a Bottle (GIAB) consortium. Version 4.2.1 is extended to include challenging medically relevant regions and other difficult to map regions. Version 4.2.1 covers 94.1% of reference (GRCh37) and v3.3.2 covers 87.8% of reference. For more information, see GIAB publication https://doi.org/10.1016/j.xgen.2022.100128.
| Sensitivity % (TP/(TP+FN) | Specificity % | |||
|---|---|---|---|---|
| GIAB Version 3.3.2 | GIAB Version 4.2.1 | GIAB Version 3.3.2 | GIAB Version 4.2.1 | |
| Single nucleotide variants | 99.57 % | 97.58 % | 100 % | 100 % |
| Insertions, deletions | ||||
| 1-10 bps | 95.38 % | 95.13 % | 100.00 % | 100.00 % |
| 11-20 bps | 99.09 % | 98.15 % | 100.00 % | 100.00 % |
| 21-50 bps | 98.78 % | 98.85 % | 100.00 % | 100.00 % |
| 2-50 bps | 97.62 % | 97.41 % | 100.00 % | 100.00 % |
| Copy number variants (exon level dels/dups, clinical sample performance) | Sensitivity | Specificity | ||
| 1 exon level deletion (heterozygous) | 100% (14/14) | NA | ||
| 1 exon deletion (homozygous or hemizygous) | 100% (5/5) | NA | ||
| 2-4 exon deletion (heterozygous or homozygous) | 100% (17/17) | NA | ||
| 5-33 exon deletion (heterozygous) | 100% (12/12) | NA | ||
| 1-5 exon duplication (heterozygous or homozygous) | 77% (10/13) | NA | ||
| 9-31 exon duplication (heterozygous) | 100% (7/7) | NA | ||
| Simulated CNV detection in reference samples (n=10) | Sensitivity | |||
| 5 exon level deletion/duplication | 98 % | |||
| Microdeletion/-duplication syndromes (large CNVs, n=22)) | ||||
| Size range (0.1-47 Mb) | 100% (22/22) | |||
| The performance presented above was reached by Blueprint Genetics high-quality, clinical grade NGS sequencing assay with the following coverage metrics | ||||
| Average of median sequencing depths in reference samples | 136x | |||
| Nucleotides with >20x sequencing coverage (%) | 99.77% | |||
Performance of Blueprint Genetics Mitochondrial Sequencing Assay.
| ANALYTIC VALIDATION (reference samples; n=4) | Sensitivity % | |||
| Single nucleotide variants | ||||
| Heteroplasmic (45-100%) | 100.0% (50/50) | |||
| Heteroplasmic (35-45%) | 100.0% (87/87) | |||
| Heteroplasmic (25-35%) | 100.0% (73/73) | |||
| Heteroplasmic (15-25%) | 100.0% (74/74) | |||
| Heteroplasmic (5-15%) | 100.0% (79/79) | |||
| Heteroplasmic (<5%) | 53.3 % (8/15) | |||
| CLINICAL VALIDATION (n=20 samples) | ||||
| Single nucleotide variants (n=18 SNVs) | 100.0% (3/3) | |||
| Heteroplasmic (10-15%) | 100.0% (5/5) | |||
| Heteroplasmic (5-10%) | 100.0% (5/5) | |||
| Heteroplasmic (<5%) | 20% (1/5) | |||
| Insertions and deletions by sequence analysis (n=3) | ||||
| Heteroplasmic (45-100%) 1-10bp | 100.0% (3/3) | |||
| Validation of the mitochondrial genome analysis workflow (based on simulated data of pathogenic mitomap mutations) | ||||
| Insertions and deletions 1-24 bps by sequence analysis; n=17 | ||||
| Homoplasmic (100%) 1-24bp | 100.0% (17/17) | |||
| Heteroplasmic (50%) | 100.0% (17/17) | |||
| Heteroplasmic (25%) | 100.0% (17/17) | |||
| Heteroplasmic (20%) | 100.0% (17/17) | |||
| Heteroplasmic (15%) | 100.0% (17/17) | |||
| Heteroplasmic (10%) | 94.1% (16/17) | |||
| Heteroplasmic (5%) | 94.1% (16/17) | |||
| Copy number variants (separate artifical mutations; n=1500) | ||||
| Homoplasmic (100%) 500 bp, 1kb, 5 kb | 100.0% | |||
| Heteroplasmic (50%) 500 bp, 1kb, 5 kb | 100.0% | |||
| Heteroplasmic (30%) 500 bp, 1kb, 5 kb | 100.0% | |||
| Heteroplasmic (20%) 500 bp, 1kb, 5 kb | 99.7% | |||
| Heteroplasmic (10%) 500 bp, 1kb, 5 kb | 99.0% | |||
| Following mtDNA coverage metrics were obtained in clinical samples in the assay validation (n=238) | ||||
| Mean of medians | ||||
| Mean sequencing depth MQ0 | 6334x | |||
| Nucleotides with >1000x MQ0 sequencing coverage (%) | 100% | |||
| rho zero cell line (=no mtDNA), mean sequencing depth in mitochondrial assay validation | 12X | |||
- Log in to our online portal Nucleus at nucleus.blueprintgenetics.com for the easiest way to order and access results.
- Fill out both the consent and requisition forms.
- Send the sample to Blueprint Genetics.
- Sample Requirements:
- Blood (min. 1ml) in an EDTA tube
- Extracted DNA, min. 2 μg in TE buffer or equivalent
- Saliva (Oragene DNA OG-500 kit/OGD-500 or OG-575 & OGD-575)
- Read more about our sample requirements
- Sending a positive control sample from the index patient is not required.
- Sample Requirements:
For samples coming to Helsinki laboratory Targeted Variant Testing is done using Next Generation Sequencing (NGS). When confirmation is required, this is done using Sanger sequencing or dPCR.
For samples coming to Marlborough laboratory Targeted Variant Testing is done with Next Generation Sequencing (NGS) technology and/or Sanger sequencing and dPCR.